Results
Contents
Results#
Workflow for enzyme kinetics parameter estimation#
In this work, a Python-based analysis workflow for kinetic parameter estimation of enzyme reactions was developed. The workflow itself consists of the steps (i) data acquisition, (ii) raw data preparation, (iii) concentration calculation, (iv) quality control, (v) modeling, and (vi) saving of results. For precise concentration calculations as well as kinetic parameter estimation the two Python modules CaliPytion and EnzymePynetics were respectively developed over the course of this work.
The two packages were designed to seamlessly interact with EnzymeML documents which were modified using PyEnzyme and thus allowing a continuous data stream from analytical raw data to kinetic parameters.
Starting from raw data of photometric measurements, which are stored in an EnzymeML document, concentration values are calculated by CaliPytion, whereupon kinetic parameters are determined by EnzymePynetics. Finally, the modeling results are written back to the EnzymeML document via PyEnzyme (Fig. 1).
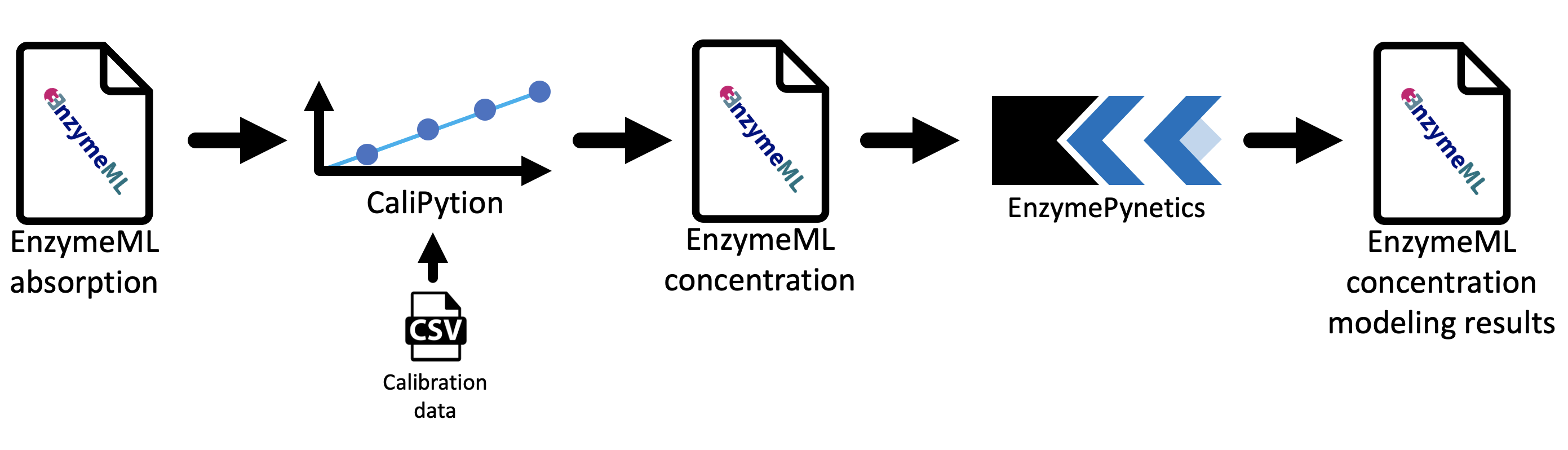 Fig. 1: Components of the kinetic parameter estimation workflow for enzyme reaction data.
Fig. 1: Components of the kinetic parameter estimation workflow for enzyme reaction data.
A key aspect of the workflow design was to enable reproducible analysis. Therefore, documentation of raw data treatment prior to modeling is vital. As a result, Jupyter Notebooks were used as a platform, since they allow to write text and code within the same document.
CaliPytion#
CaliPytion was developed to facilitate robust conversion of measurement data to concentration data. Based on an analyte standard, a calibration curve is created. Since linear and non-linear relations between analyte concentration and analytic signal may occur, (Hsu and Chen, 2010), (Martin et al., 2017), linear, polynomial, exponential, and rational calibration equations were implemented. After fitting, the best model is preselected based on Akaike information criterion (Akaike, 1998), offering a metric to compare models with different number of parameters. CaliPytion was designed to work with PyEnzyme. Hence, a StandardCurve of CaliPytion can natively by applied to an EnzymeMLDocument of PyEnzyme. As a result, absorption data of an EnzymeML document can be converted into concentration data based on the previously generated standard curve.
EnzymePynetics#
EnzymePynetics was developed to enable easy applicable parameter estimation of single substrate enzyme reactions directly from EnzymeML documents. At its core, the ParameterEstimator of EnzymePynetics fits experimental data to different Michaels-Menten type models. Besides the irreversible Michaelis-Menten model, competitive, non-competitive, uncompetitive, and partially competitive inhibition models were implemented. Hence, the inhibition constant of potentially inhibiting substrate or product can be estimated. Furthermore, the inhibition constant of an applied inhibitor can be quantified. Since, the experimental data is fitted against multiple kinetic models, the best fitting model is suggested based on AIC. Hence, a proposal of the enzyme’s kinetic mechanism is given based on the best fitting model.
For quality control, visualization of experimental data as well as the fitted kinetic model was a priority during development. Thereby, detection of systematic deviation between model and measurement data is possible. Furthermore, the ParameterEstimator allows to subset the measurement data by time as well as initial substrate concentration without deleting data. Hence, identified systematic deviations (e.g. lag-phases or measurements with incorrect enzyme concentrations) can be excluded from parameter estimation.
Like the StandardCurve of CaliPytion, the ParameterEstimator of EnzymePynetics natively supports EnzymeMLDocuments as a data source.
Scenario-driven workflow development#
The development of the workflow was driven by different research scenarios of EnzymeML project partners. The goal of all projects was to reliably determine kinetic parameters of enzyme reactions.
The collaboration consisted of multiple rounds of lab experiments (preformed by the experimental partners) and kinetic modeling of experimental data. Each round included (i) wet lab experiments, (ii) kinetic modeling, (iii) design of follow-up experiments, and (iv) discussion of results with the respective project partners. Hence, a short feedback loop between lab experiments and modeling-based experimental suggestions was established.
In parallel, the Python modules CaliPytion and EnzymePynetics were developed. Thereby, individual requirements of each research scenario fostered the implementation of various functionalities, resulting in a feature-rich yet generic workflow.
Data analysis with Jupyter Notebooks#
All data analysis was carried out in Jupyter Notebooks by applying the developed workflow for kinetic parameter estimation. Within the next chapters, the latest results of all four experimental scenarios are shown. Thereby, the applicability of the workflow is demonstrated while analyzing and discussing the results of the respective scenario. Since the analysis was conducted in Jupyter Notebooks, each of the following chapter is a executed Jupyter Notebook. Hence all figures and tables for data visualization were generated at runtime of the analysis. Thereby, the applicability of Jupyter Notebooks for reproducible and comprehensive computational workflows in a scientific environment is demonstrated. Every scenario consists of a short description of the project’s background and methodology carried out by the project partners in the laboratory. Additionally, data preparation, kinetic modeling steps, and the results are shown was well as project specific results are discussed. All notebooks can be launched interactively by clicking on the -icon in the upper section of each scenario page. As a result, all analysis can be interactively repeated. Therefore, it is highly recommended to read this work in its conceptualized form.